SHAMSUL Demo App: https://shamsul.serve.scilifelab.se/
SHAMSUL
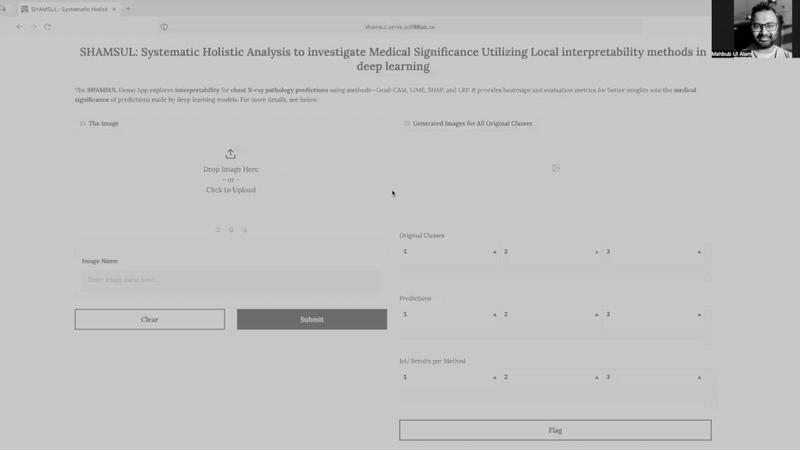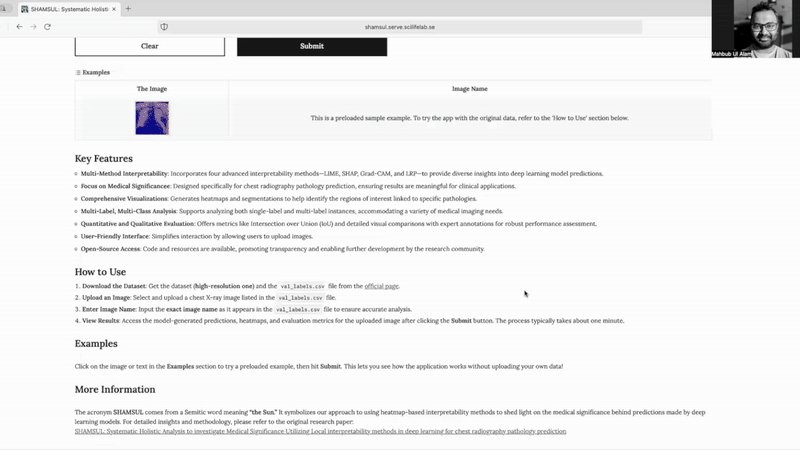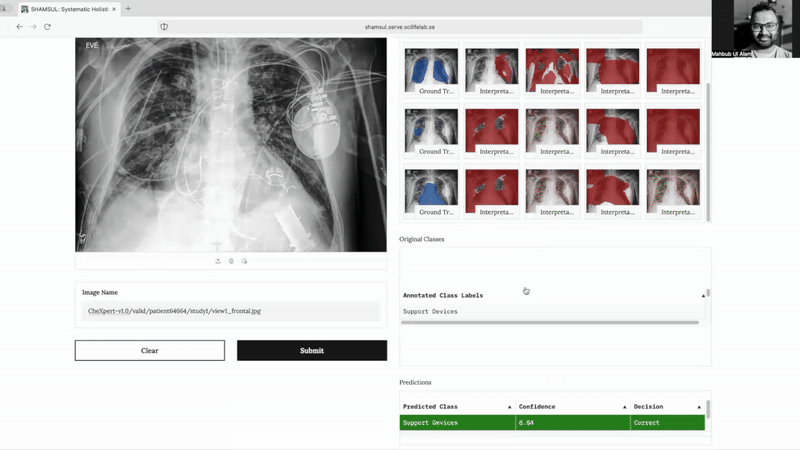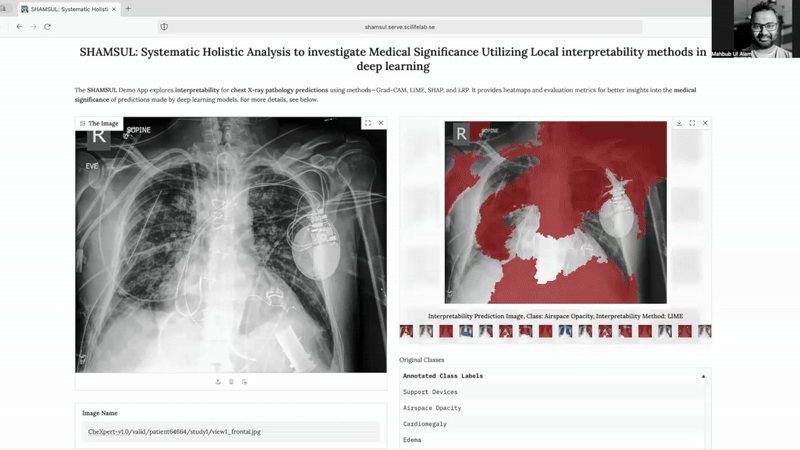
SHAMSUL* explores interpretability for chest X-ray pathology predictions using methods—Grad-CAM, LIME, SHAP, and LRP. It provides heatmaps and evaluation metrics for better insights into the medical significance of predictions made by deep learning models.
For detailed insights and methodology, please refer to the original research paper:
SHAMSUL: Systematic Holistic Analysis to investigate Medical Significance Utilizing Local interpretability methods in deep learning for chest radiography pathology prediction
*”The acronym SHAMSUL, derived from a Semitic word meaning “the Sun,” serves as a symbolic representation of our heatmap score-based interpretability analysis approach aimed at unveiling the medical significance inherent in the predictions of black box deep learning models.”
Demo App- Visit https://shamsul.serve.scilifelab.se/
Explore this work using the publicly available demo app (no registration needed!):
Online Access
Visit https://shamsul.serve.scilifelab.se/ to use it directly.
OR
Run Locally
Step 1: Install Docker
Install Docker Engine or Docker Desktop on your system by following the official Docker installation guide.
Step 2: Launch the App
2a. Open a Terminal (or Windows Terminal).
2b. Run this command to download and start the app:
docker run --rm --name shamsul -p 7860:7860 mahbub1969/shamsul:v6
2c. Open your browser and go to http://localhost:7860/ to use the app.
Step 3: Stop the App
To stop the app, press Control+C in the terminal. Note that the session won’t be saved, so the app will reset to its default state the next time you run it.
Step 4: Remove the Docker Image (Optional)
If you want to free up space, you can remove the Docker image. Use this command in your terminal:
docker image rm mahbub1969/shamsul:v6
For more details, check out the Docker image removal guide.
Key Features
- Multi-Method Interpretability: Incorporates four advanced interpretability methods—LIME, SHAP, Grad-CAM, and LRP—to provide diverse insights into deep learning model predictions.
- Focus on Medical Significance: Designed specifically for chest radiography pathology prediction, ensuring results are meaningful for clinical applications.
- Comprehensive Visualizations: Generates heatmaps and segmentations to help identify the regions of interest linked to specific pathologies.
- Multi-Label, Multi-Class Analysis: Supports analyzing both single-label and multi-label instances, accommodating a variety of medical imaging needs.
- Quantitative and Qualitative Evaluation: Offers metrics like Intersection over Union (IoU) and detailed visual comparisons with expert annotations for robust performance assessment.
- User-Friendly Interface: Simplifies interaction by allowing users to upload images.
- Open-Source Access: Code and resources are available, promoting transparency and enabling further development by the research community.
An excerpt of the SHAMSUL paper
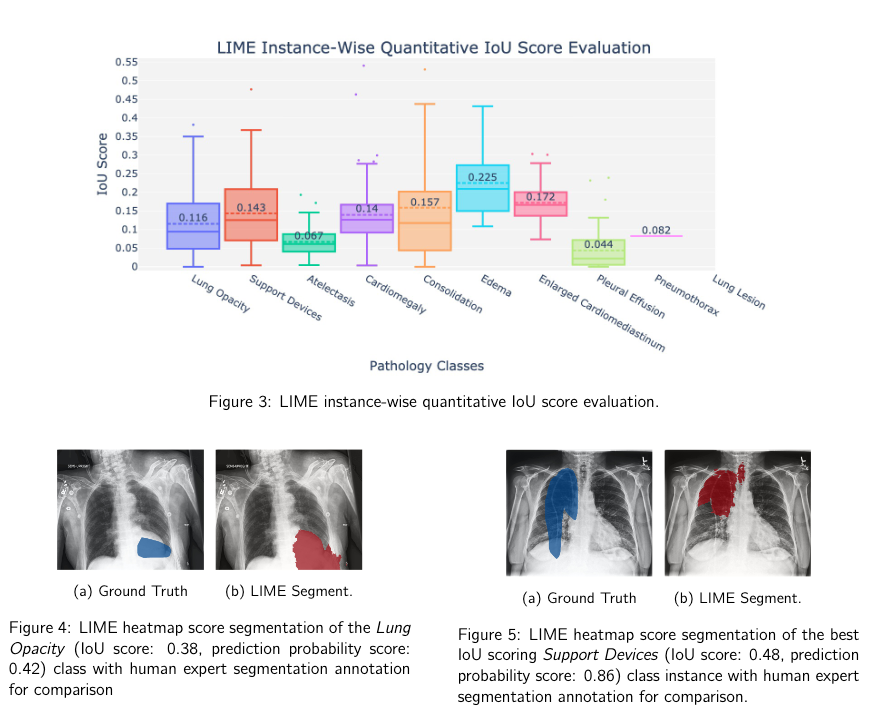
Citation
Please acknowledge the following work in papers or derivative software:
M. U. Alam, J. Hollmén, J. R. Baldvinsson, and R. Rahmani, “SHAMSUL: Systematic Holistic Analysis to investigate Medical Significance Utilizing Local interpretability methods in deep learning for chest radiography pathology prediction,” Nordic Machine Intelligence, vol. 3, pp. 27–47, 2023. https://doi.org/10.5617/nmi.10471
Bibtex Format for Citation
@article{alam2023shamsul,
title={SHAMSUL: Systematic Holistic Analysis to investigate Medical Significance Utilizing Local interpretability methods in deep learning for chest radiography pathology prediction},
author={Ul Alam, Mahbub and Hollmén, Jaakko and Baldvinsson, Jón Rúnar and Rahmani, Rahim},
journal={Nordic Machine Intelligence},
volume={3},
number={1},
pages={27--47},
year={2023},
doi={10.5617/nmi.10471}
}